A pre-trained model for Nuclei Classification within Haematoxylin & Eosin stained histology images
A pre-trained model for classifying nuclei cells as the following types - Other - Inflammatory - Epithelial - Spindle-Shaped
This model is trained using DenseNet121 over ConSeP dataset.
Data
The training dataset is from https://warwick.ac.uk/fac/cross _fac/tia/data/hovernet
wget https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip
unzip -q consep_dataset.zip
Preprocessing
After downloading this dataset
, python script data_process.py from scripts folder can be used to preprocess and generate the final dataset for training.
python scripts/data_process.py --input /path/to/data/CoNSeP --output /path/to/data/CoNSePNuclei
After generating the output files, please modify the dataset_dir parameter specified in configs/train.json and configs/inference.json to reflect the output folder which contains new dataset.json.
Class values in dataset are
- 1 = other
- 2 = inflammatory
- 3 = healthy epithelial
- 4 = dysplastic/malignant epithelial
- 5 = fibroblast
- 6 = muscle
- 7 = endothelial
As part of pre-processing, the following steps are executed.
- Crop and Extract each nuclei Image + Label (128x128) based on the centroid given in the dataset.
- Combine classes 3 & 4 into the epithelial class and 5,6 & 7 into the spindle-shaped class.
- Update the label index for the target nuclie based on the class value
- Other cells which are part of the patch are modified to have label idex = 255
Example dataset.json in output folder:
{
"training": [
{
"image": "/workspace/data/CoNSePNuclei/Train/Images/train_1_3_0001.png",
"label": "/workspace/data/CoNSePNuclei/Train/Labels/train_1_3_0001.png",
"nuclei_id": 1,
"mask_value": 3,
"centroid": [
64,
64
]
}
],
"validation": [
{
"image": "/workspace/data/CoNSePNuclei/Test/Images/test_1_3_0001.png",
"label": "/workspace/data/CoNSePNuclei/Test/Labels/test_1_3_0001.png",
"nuclei_id": 1,
"mask_value": 3,
"centroid": [
64,
64
]
}
]
}
Training configuration
The training was performed with the following:
- GPU: at least 12GB of GPU memory
- Actual Model Input: 4 x 128 x 128
- AMP: True
- Optimizer: Adam
- Learning Rate: 1e-4
- Loss: torch.nn.CrossEntropyLoss
- Dataset Manager: CacheDataset
Memory Consumption Warning
If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate cache_rate in the configurations within range [0, 1] to minimize the System RAM requirements.
Input
4 channels - 3 RGB channels - 1 signal channel (label mask)
Output
4 channels - 0 = Other - 1 = Inflammatory - 2 = Epithelial - 3 = Spindle-Shaped
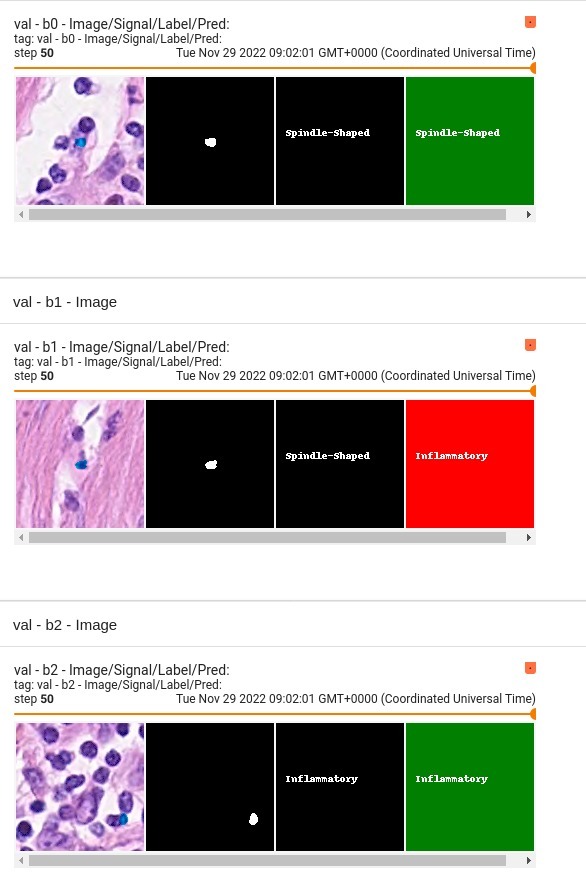
Performance
This model achieves the following F1 score on the validation data provided as part of the dataset:
- Train F1 score = 0.926
- Validation F1 score = 0.852
Confusion Metrics for Validation for individual classes are:
Metric
Other
Inflammatory
Epithelial
Spindle-Shaped
Precision
0.6909
0.7773
0.9078
0.8478
Recall
0.2754
0.7831
0.9533
0.8514
F1-score
0.3938
0.7802
0.9300
0.8496
Confusion Metrics for Training for individual classes are:
Metric
Other
Inflammatory
Epithelial
Spindle-Shaped
Precision
0.8000
0.9076
0.9560
0.9019
Recall
0.6512
0.9028
0.9690
0.8989
F1-score
0.7179
0.9052
0.9625
0.9004
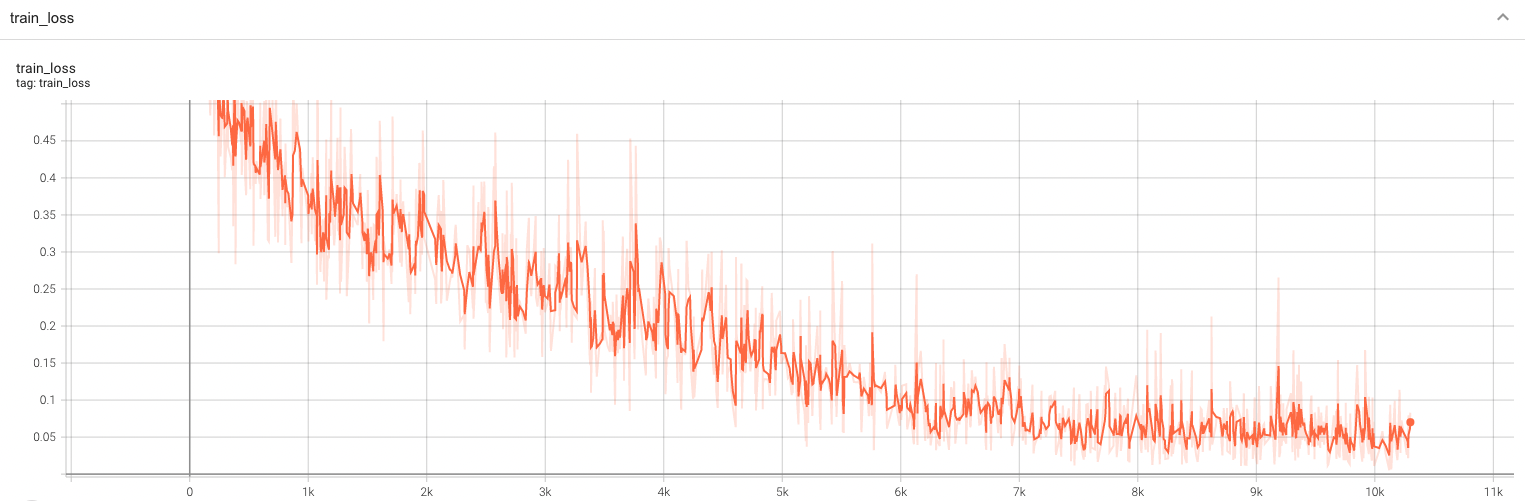
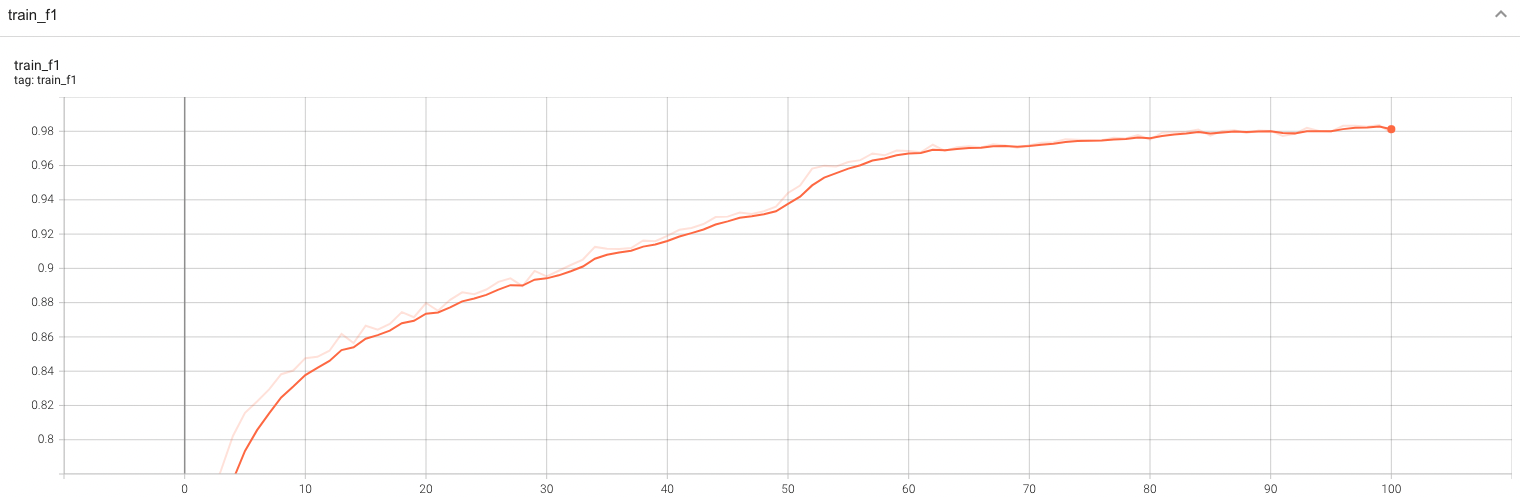
Training Loss and F1
A graph showing the training Loss and F1-score over 100 epochs.
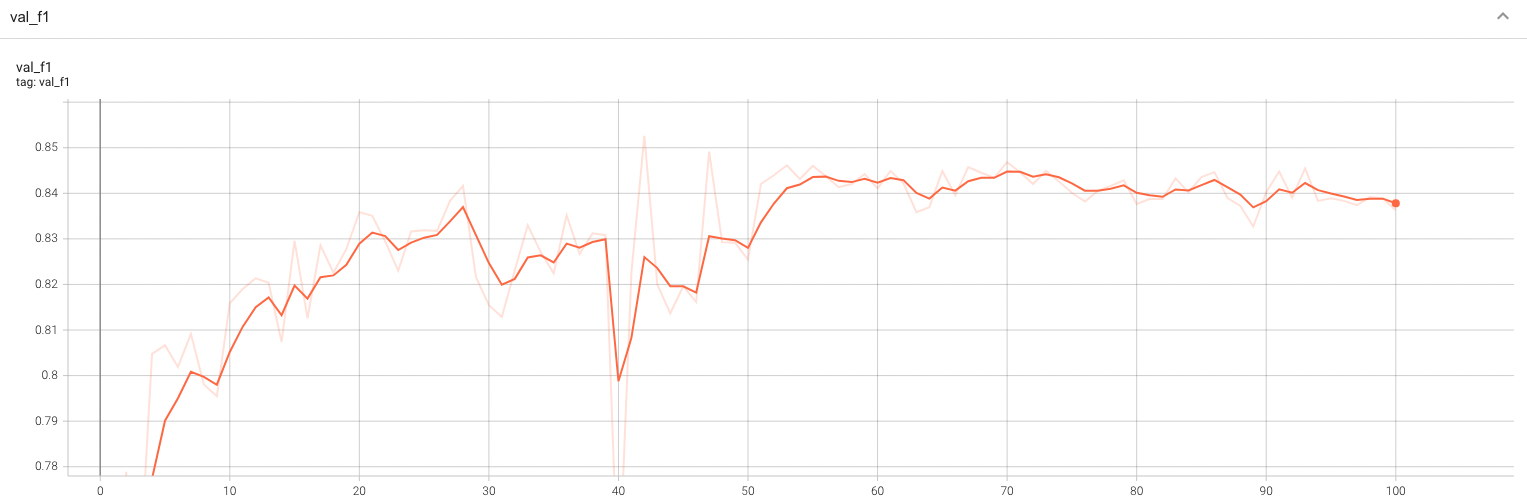
Validation F1
A graph showing the validation F1-score over 100 epochs.
MONAI Bundle Commands
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
For more details usage instructions, visit the MONAI Bundle Configuration Page .
Execute training:
python -m monai.bundle run --config_file configs/train.json
Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using --dataset_dir :
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
Override the train config to execute multi-GPU training:
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove --standalone , modify --nnodes , or do some other necessary changes according to the machine used. For more details, please refer to pytorch’s official tutorial
.
Override the train config to execute evaluation with the trained model:
python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
Override the train config and evaluate config to execute multi-GPU evaluation:
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json','configs/multi_gpu_evaluate.json']"
Execute inference:
python -m monai.bundle run --config_file configs/inference.json
References
[1] S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. “HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images.” Medical Image Analysis, Sept. 2019. [ doi ]
License
Copyright (c) MONAI Consortium
Licensed under the Apache License, Version 2.0 (the “License”); you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an “AS IS” BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
Follow AI Models on Google News
An easy & free way to support AI Models is to follow our google news feed! More followers will help us reach a wider audience!
Google News: AI Models