A 3D transformer-based model for whole brain segmentation from T1W MRI image
Detailed whole brain segmentation is an essential quantitative technique in medical image analysis, which provides a non-invasive way of measuring brain regions from a clinical acquired structural magnetic resonance imaging (MRI). We provide the pre-trained model for training and inferencing whole brain segmentation with 133 structures. Training pipeline is provided to support active learning in MONAI Label and training with bundle.
A tutorial and release of model for whole brain segmentation using the 3D transformer-based segmentation model UNEST.
Authors: Xin Yu (xin.yu@vanderbilt.edu )
Yinchi Zhou (yinchi.zhou@vanderbilt.edu ) | Yucheng Tang (yuchengt@nvidia.com )
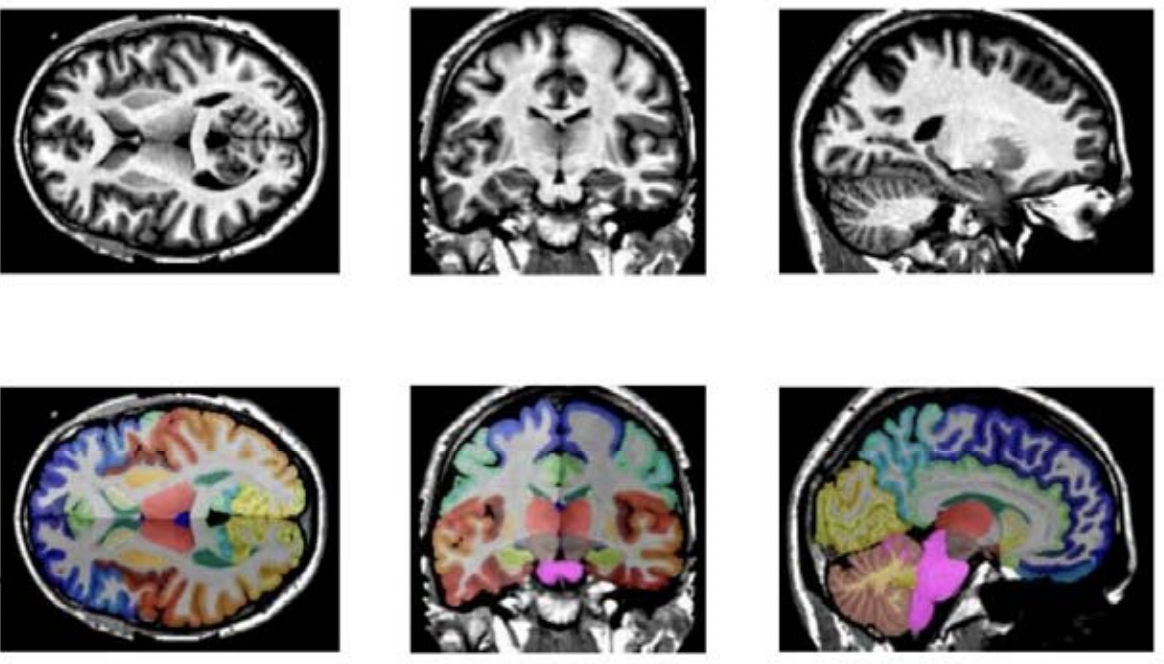
Fig.1 - The demonstration of T1w MRI images registered in MNI space and the whole brain segmentation labels with 133 classes
Model Overview
A pre-trained UNEST base model [1] for volumetric (3D) whole brain segmentation with T1w MR images. To leverage information across embedded sequences, ”shifted window” transformers are proposed for dense predictions and modeling multi-scale features. However, these attempts that aim to complicate the self-attention range often yield high computation complexity and data inefficiency. Inspired by the aggregation function in the nested ViT, we propose a new design of a 3D U-shape medical segmentation model with Nested Transformers (UNesT) hierarchically with the 3D block aggregation function, that learn locality behaviors for small structures or small dataset. This design retains the original global self-attention mechanism and achieves information communication across patches by stacking transformer encoders hierarchically.
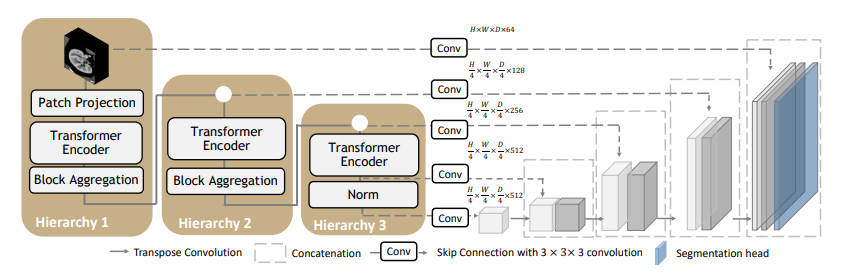
Fig.2 - The network architecture of UNEST Base model
Data
The training data is from the Vanderbilt University and Vanderbilt University Medical Center with public released OASIS and CANDI datsets. Training and testing data are MRI T1-weighted (T1w) 3D volumes coming from 3 different sites. There are a total of 133 classes in the whole brain segmentation task. Among 50 T1w MRI scans from Open Access Series on Imaging Studies (OASIS) (Marcus et al., 2007) dataset, 45 scans are used for training and the other 5 for validation. The testing cohort contains Colin27 T1w scan (Aubert-Broche et al., 2006) and 13 T1w MRI scans from the Child and Adolescent Neuro Development Initiative (CANDI) (Kennedy et al., 2012). All data are registered to the MNI space using the MNI305 (Evans et al., 1993) template and preprocessed follow the method in (Huo et al., 2019). Input images are randomly cropped to the size of 96 × 96 × 96.
Important
The brain MRI images for training are registered to Affine registration from the target image to the MNI305 template using NiftyReg. The data should be in the MNI305 space before inference.
If your images are already in MNI space, skip the registration step.
You could use any resitration tool to register image to MNI space. Here is an example using ants. Registration to MNI Space: Sample suggestion. E.g., use ANTS or other tools for registering T1 MRI image to MNI305 Space.
pip install antspyx
#Sample ANTS registration
import ants
import sys
import os
fixed_image = ants.image_read('<fixed_image_path>')
moving_image = ants.image_read('<moving_image_path>')
transform = ants.registration(fixed_image,moving_image,'Affine')
reg3t = ants.apply_transforms(fixed_image,moving_image,transform['fwdtransforms'][0])
ants.image_write(reg3t,output_image_path)
Training configuration
The training and inference was performed with at least one 24GB-memory GPU.
Actual Model Input: 96 x 96 x 96
Input and output formats
Input: 1 channel T1w MRI image in MNI305 Space.
commands example
Download trained checkpoint model to ./model/model.pt:
Add scripts component: To run the workflow with customized components, PYTHONPATH should be revised to include the path to the customized component:
export PYTHONPATH=$PYTHONPATH: '<path to the bundle root dir>/scripts'
Execute Training:
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
Execute inference:
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
More examples output
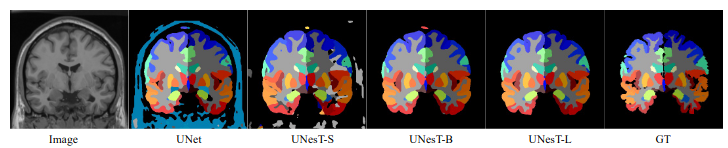
Fig.3 - The output prediction comparison with variant and ground truth
Training/Validation Benchmarking
A graph showing the training accuracy for fine-tuning 600 epochs.
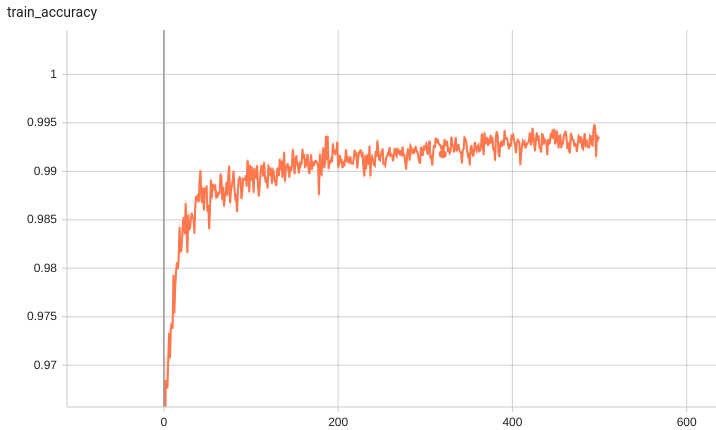
With 10 fine-tuned labels, the training process converges fast.
Complete ROI of the whole brain segmentation
133 brain structures are segmented.
#1
#2
#3
#4
0: background
1 : 3rd-Ventricle
2 : 4th-Ventricle
3 : Right-Accumbens-Area
4 : Left-Accumbens-Area
5 : Right-Amygdala
6 : Left-Amygdala
7 : Brain-Stem
8 : Right-Caudate
9 : Left-Caudate
10 : Right-Cerebellum-Exterior
11 : Left-Cerebellum-Exterior
12 : Right-Cerebellum-White-Matter
13 : Left-Cerebellum-White-Matter
14 : Right-Cerebral-White-Matter
15 : Left-Cerebral-White-Matter
16 : Right-Hippocampus
17 : Left-Hippocampus
18 : Right-Inf-Lat-Vent
19 : Left-Inf-Lat-Vent
20 : Right-Lateral-Ventricle
21 : Left-Lateral-Ventricle
22 : Right-Pallidum
23 : Left-Pallidum
24 : Right-Putamen
25 : Left-Putamen
26 : Right-Thalamus-Proper
27 : Left-Thalamus-Proper
28 : Right-Ventral-DC
29 : Left-Ventral-DC
30 : Cerebellar-Vermal-Lobules-I-V
31 : Cerebellar-Vermal-Lobules-VI-VII
32 : Cerebellar-Vermal-Lobules-VIII-X
33 : Left-Basal-Forebrain
34 : Right-Basal-Forebrain
35 : Right-ACgG–anterior-cingulate-gyrus
36 : Left-ACgG–anterior-cingulate-gyrus
37 : Right-AIns–anterior-insula
38 : Left-AIns–anterior-insula
39 : Right-AOrG–anterior-orbital-gyrus
40 : Left-AOrG–anterior-orbital-gyrus
41 : Right-AnG—angular-gyrus
42 : Left-AnG—angular-gyrus
43 : Right-Calc–calcarine-cortex
44 : Left-Calc–calcarine-cortex
45 : Right-CO—-central-operculum
46 : Left-CO—-central-operculum
47 : Right-Cun—cuneus
48 : Left-Cun—cuneus
49 : Right-Ent—entorhinal-area
50 : Left-Ent—entorhinal-area
51 : Right-FO—-frontal-operculum
52 : Left-FO—-frontal-operculum
53 : Right-FRP—frontal-pole
54 : Left-FRP—frontal-pole
55 : Right-FuG—fusiform-gyrus
56 : Left-FuG—fusiform-gyrus
57 : Right-GRe—gyrus-rectus
58 : Left-GRe—gyrus-rectus
59 : Right-IOG—inferior-occipital-gyrus ,
60 : Left-IOG—inferior-occipital-gyrus
61 : Right-ITG—inferior-temporal-gyrus
62 : Left-ITG—inferior-temporal-gyrus
63 : Right-LiG—lingual-gyrus
64 : Left-LiG—lingual-gyrus
65 : Right-LOrG–lateral-orbital-gyrus
66 : Left-LOrG–lateral-orbital-gyrus
67 : Right-MCgG–middle-cingulate-gyrus
68 : Left-MCgG–middle-cingulate-gyrus
69 : Right-MFC—medial-frontal-cortex
70 : Left-MFC—medial-frontal-cortex
71 : Right-MFG—middle-frontal-gyrus
72 : Left-MFG—middle-frontal-gyrus
73 : Right-MOG—middle-occipital-gyrus
74 : Left-MOG—middle-occipital-gyrus
75 : Right-MOrG–medial-orbital-gyrus
76 : Left-MOrG–medial-orbital-gyrus
77 : Right-MPoG–postcentral-gyrus
78 : Left-MPoG–postcentral-gyrus
79 : Right-MPrG–precentral-gyrus
80 : Left-MPrG–precentral-gyrus
81 : Right-MSFG–superior-frontal-gyrus
82 : Left-MSFG–superior-frontal-gyrus
83 : Right-MTG—middle-temporal-gyrus
84 : Left-MTG—middle-temporal-gyrus
85 : Right-OCP—occipital-pole
86 : Left-OCP—occipital-pole
87 : Right-OFuG–occipital-fusiform-gyrus
88 : Left-OFuG–occipital-fusiform-gyrus
89 : Right-OpIFG-opercular-part-of-the-IFG
90 : Left-OpIFG-opercular-part-of-the-IFG
91 : Right-OrIFG-orbital-part-of-the-IFG
92 : Left-OrIFG-orbital-part-of-the-IFG
93 : Right-PCgG–posterior-cingulate-gyrus
94 : Left-PCgG–posterior-cingulate-gyrus
95 : Right-PCu—precuneus
96 : Left-PCu—precuneus
97 : Right-PHG—parahippocampal-gyrus
98 : Left-PHG—parahippocampal-gyrus
99 : Right-PIns–posterior-insula
100 : Left-PIns–posterior-insula
101 : Right-PO—-parietal-operculum
102 : Left-PO—-parietal-operculum
103 : Right-PoG—postcentral-gyrus
104 : Left-PoG—postcentral-gyrus
105 : Right-POrG–posterior-orbital-gyrus
106 : Left-POrG–posterior-orbital-gyrus
107 : Right-PP—-planum-polare
108 : Left-PP—-planum-polare
109 : Right-PrG—precentral-gyrus
110 : Left-PrG—precentral-gyrus
111 : Right-PT—-planum-temporale
112 : Left-PT—-planum-temporale
113 : Right-SCA—subcallosal-area
114 : Left-SCA—subcallosal-area
115 : Right-SFG—superior-frontal-gyrus
116 : Left-SFG—superior-frontal-gyrus
117 : Right-SMC—supplementary-motor-cortex
118 : Left-SMC—supplementary-motor-cortex
119 : Right-SMG—supramarginal-gyrus
120 : Left-SMG—supramarginal-gyrus
121 : Right-SOG—superior-occipital-gyrus
122 : Left-SOG—superior-occipital-gyrus
123 : Right-SPL—superior-parietal-lobule
124 : Left-SPL—superior-parietal-lobule
125 : Right-STG—superior-temporal-gyrus
126 : Left-STG—superior-temporal-gyrus
127 : Right-TMP—temporal-pole
128 : Left-TMP—temporal-pole
129 : Right-TrIFG-triangular-part-of-the-IFG
130 : Left-TrIFG-triangular-part-of-the-IFG
131 : Right-TTG—transverse-temporal-gyrus
132 : Left-TTG—transverse-temporal-gyrus
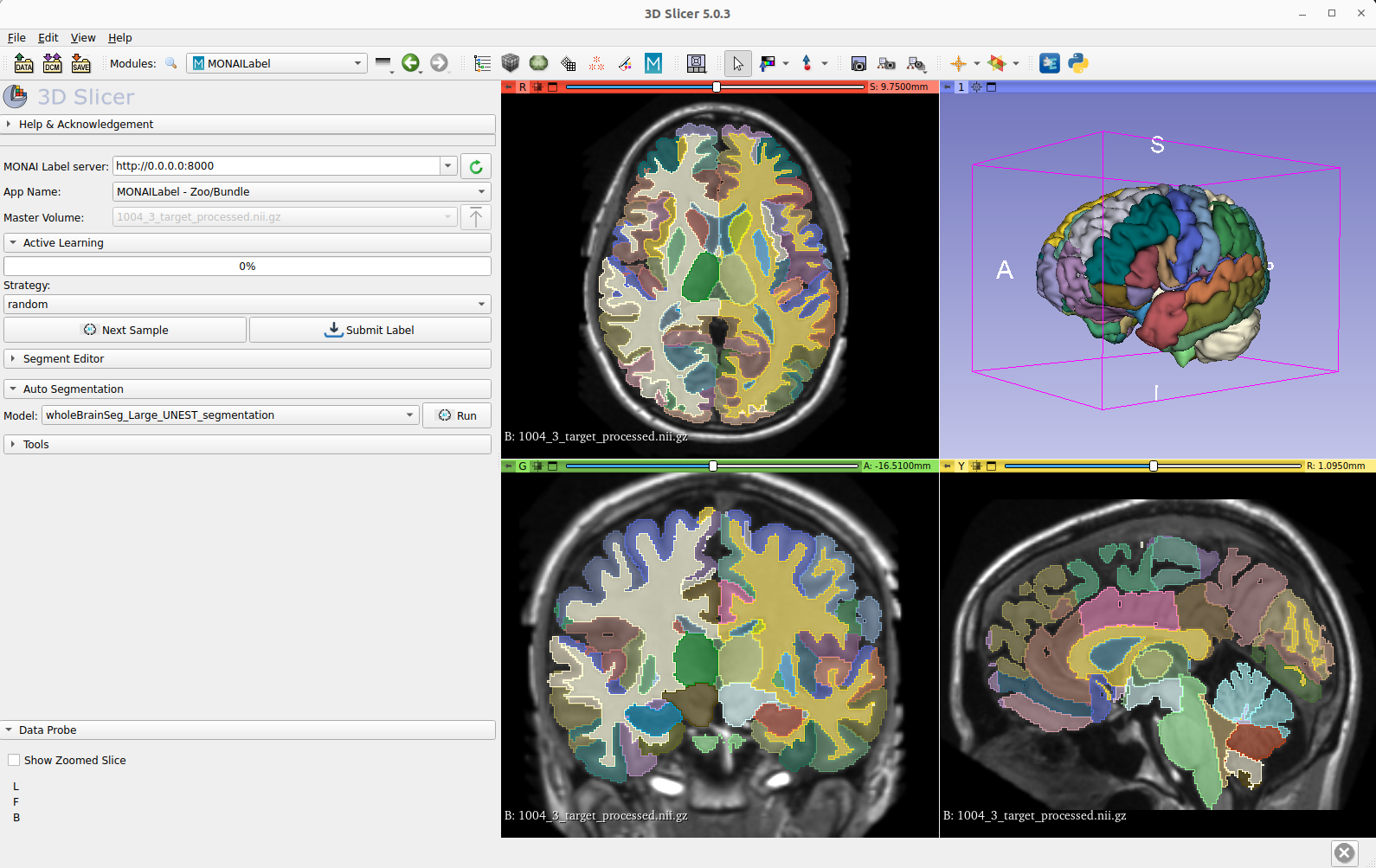
Bundle Integration in MONAI Lable
The inference and training pipleine can be easily used by the MONAI Label server and 3D Slicer for fast labeling T1w MRI images in MNI space.
Disclaimer
This is an example, not to be used for diagnostic purposes.
References
[1] Yu, Xin, Yinchi Zhou, Yucheng Tang et al. Characterizing Renal Structures with 3D Block Aggregate Transformers. arXiv preprint arXiv:2203.02430 (2022). https://arxiv.org/pdf/2203.02430.pdf
[2] Zizhao Zhang et al. Nested Hierarchical Transformer: Towards Accurate, Data-Efficient and Interpretable Visual Understanding. AAAI Conference on Artificial Intelligence (AAAI) 2022
[3] Huo, Yuankai, et al. 3D whole brain segmentation using spatially localized atlas network tiles. NeuroImage 194 (2019): 105-119.
License
Copyright (c) MONAI Consortium
Licensed under the Apache License, Version 2.0 (the “License”); you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an “AS IS” BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
Follow AI Models on Google News
An easy & free way to support AI Models is to follow our google news feed! More followers will help us reach a wider audience!
Google News: AI Models


